Abstract
Objective: Incidence rates of colon and liver cancers differ dramatically between Northern vs. Southern India. It has been suggested that differences in regional diet may play a role, specifically in the disparity in consumption rates of curcumin, an active agent of turmeric, and epigallocatechin gallate (EGCG), a compound in green tea. Curcumin and EGCG have well-known multi-targeted and beneficial effects as chemopreventive agents. However, natural compounds typically require high concentrations to be effective, which can also negatively impact healthy cells. Alternatively, low-dose combination of these compounds, if proven effective, may be one way to avoid this problem. This study proposed to demonstrate the effects of individual and combined treatments of EGCG and curcumin on viability and cancer pathway signaling, in hepatocellular carcinoma (HCC) and colorectal cancer (CCR) cell lines.
Methods: HCC and CCR cell lines (HepG2 and SW1417, respectively) were treated with curcumin and EGCG in a dose- and time-dependent manner. Regorafenib, a chemotherapeutic used to treat colon and liver cancers, was then combined with the effective low-dose EGCG/curcumin. Cell viability, proliferation, and expression of cancer target genes were assessed.
Results: Low-dose combination of curcumin and EGCG influenced the expression of 28 cancer target genes in HepG2 and 14 genes in SW1417. Six of these targets were verified by quantitative polymerase chain reaction. Regorafenib treatment elicited an effect similar to curcumin + EGCG treatment in a number of these targets and enhanced their regulation when used in combination.
Conclusions: Low-dose combinations of curcumin and EGCG have a beneficial effect on regulating important cancer targets in HepG2 and SW1417 cell lines. The data suggest a supportive role for phytochemicals as complementary treatments to chemotherapy.
Abbreviations
- ANGPT2
- angiopoietin 2
- ARNT
- aryl hydrocarbon receptor nuclear translocator
- ARPE-19
- adult retinal pigmented epithelium
- CRC
- colorectal cancer
- EGCG
- epigallocatechin-3-gallate
- EGF
- epithelial growth factor
- FGF2
- fibroblast growth factor 2
- HCC
- hepatocellular carcinoma
- HepG2
- hepatocellular carcinoma cell line
- IGF
- insulin-like growth factor
- IGFBP7
- insulin-like growth factor–binding protein 7
- SERPINF1
- serpin family, member 1
- SW1417
- colorectal cancer cell line
- VEGF
- vascular endothelial growth factor
INTRODUCTION
Colorectal cancer (CRC) ranks fourth among the most commonly diagnosed cancers as well as the fourth highest cause of cancer mortality among both men and women in the USA. Hepatocellular carcinoma (HCC) ranks fifteenth among cancer diagnoses, but is the fifth major cause of cancer deaths in males and females in the USA. 1 Interestingly, the incidence of colon cancer is much lower in India compared to the USA; however, the incidence of liver cancer is higher. 2 HCC is the result of a complex combination of genetic and epigenetic alterations and has the fifth highest mortality rate of all cancers reported.
Genomic studies of HCC suggest that the most frequently dysregulated pathways are those involving cell proliferation, migration, and angiogenesis. The most important of these pathways include growth factor signaling, such as insulin-like growth factor (IGF), epidermal growth factor (EGF), and vascular endothelial growth factor (VEGF). 3 Colorectal carcinoma has a very complex pathogenesis and is influenced by a variety of factors including diet, lifestyle, and genetic predisposition. 4 There are numerous pathways involved in the initiation of neoplastic transformation of CRC leading to metastasis and progression. Principle mutations leading to initiation and progression of CRC include the chromosomal instability pathway (CIN), the DNA mismatch repair (MMR) system, inflammatory pathways, and abnormal DNA methylation. 5 , 6
It has been estimated that more than half of cancer patients incorporate dietary modifications and the use of chemopreventive compounds, either exclusively or in conjunction with traditional chemotherapeutic regimens. 7 Chemoprevention typically refers to the use of either naturally occurring or synthetic compounds to inhibit or control cancer. 8 Polyphenols are a family of compounds which are not only potential cancer therapeutics, but also excellent agents for chemoprevention. These compounds are widely known for their antioxidant properties. However, recent studies have shown that they can interact with proteins, enzymes, receptors, and transcription factors, thereby directly mediating physiological changes at the molecular level. 9 A number of polyphenols have the ability to alter cell cycle regulation, thereby inhibiting proliferation and inducing apoptosis making it a potential candidate for chemoprevention and therapy. 10
Curcumin is a hydrophobic polyphenol isolated from Curcuma longa . In traditional Ayurveda medicine, it has been used to treat a range of pathologies including ulcers, arthritis, psoriasis, jaundice, and cancer. 11 Curcumin is capable of interacting with and regulating multiple molecular targets such as growth factors, transcription factors, kinases, cytokines, and apoptosis-related proteins. 12 It binds directly with more than 30 different types of proteins including DNA polymerases, focal adhesion kinases (FAK), and protein kinase C (PKC). 13 Curcumin can also affect the expression of transcription factors by activating or inhibiting them, depending on their targets. 14
Curcumin inhibits the expression of activated protein 1 (AP1) and nuclear factor (NF)-κB, both of which are important transcription factors that influence the expression of genes involved in inflammation, apoptosis, oncogenesis, and lymphoid differentiation. 15 It also inhibits the expression of signal transducer and activator of transcription (STAT) proteins, increased levels of which result in angiogenesis. 16 Curcumin also inhibits the activity of B-catenin, which plays a crucial role in cell–cell adhesion, the malfunctioning of which leads to tumorigenesis and metastasis. 17 Several transcription factors including aryl hydrocarbon receptor (ArH), important for cell proliferation, gene regulation, and inflammation, are activated by curcumin. 18 It also plays an important role in inhibiting angiogenesis by controlling the regulation of the growth factors such as VEGF, fibroblast growth factor (FGF), and EGF. 19 Finally, curcumin exhibits anti-apoptotic and anti-proliferative effects by regulating additional growth factors which control the timing and progression of the cell cycle. 20 When these molecules are unregulated, they lead to malignancy by inducing constitutive cell proliferation.
Epigallocatechin-3-gallate (EGCG) is one of the major components of green tea, constituting approximately 50%–80% of the plant’s active agents. EGCG has been shown to have beneficial effects against cancer, diabetes, stroke, and obesity. Its potency as an antioxidant has been demonstrated to suppress the inflammatory process, which is the leading cause for transformation due to carcinogenesis. 21 EGCG has the ability to inhibit tumor progression by blocking the active site of receptors in target cells. It is also known to alter the expression of the receptors themselves. Further evidence suggests that EGCG can bind directly to carcinogens and inactivate them, hence its potential in cancer prevention. 22
EGCG has the ability to promote cell growth arrest, activate effector caspases, and downregulate NF-κB, making it a potential pro-apoptotic agent. 23 EGCG also inhibits the activity of important cell cycle promoters CDK2, CDK4, and CDK6. 24 In short, both curcumin and EGCG are potent effectors of cellular responses. This study sought to demonstrate their potential for chemoprevention and therapy when administered in low-dose combinations.
Regorafenib is a relatively new targeted chemotherapeutic for the treatment of HCC and CRC. It acts as a tyrosine kinase inhibitor to block activation of EGF and VEGF receptors, thus limiting their ability to promote cellular replication and neovascularization. 25 , 26 This study sought to demonstrate the effect of low-dose treatment of curcumin and EGCG on HCC and CRC cell lines compared to regorafenib treatment. In addition, we sought to determine whether this combination of natural compounds would enhance the effect of regorafenib in these cell lines.
MATERIALS AND METHODS
CELL CULTURE
Adult retinal pigmented epithelial cell line-19 (ARPE-19), HepG2, and SW1417 cells were purchased commercially from ATCC. ARPE-19 cells were selected as a neutral normal control because normal colon and liver cells were not both available as immortalized lines. ARPE-19 cells were derived from a 19-year-old male who died of head trauma, were not immortalized, and had not been passaged more than five times. HepG2 (HCC) cells were derived from a 15-year-old male patient, and SW1417 (CRC) cells were derived from a 53-year-old female Caucasian patient.
All cell lines were maintained using standard cell culture technique, in Dulbecco’s Modified Eagle Medium (DMEM): F-12, 1:1, supplemented with 1.0% Pen-Strep, 1.0% L-glutamine, and 10% fetal bovine serum (FBS) (HyClone). Curcumin, EGCG, and regorafenib (Sigma-Aldrich) were purchased commercially and then suspended at 1.0 mM in a complete culture medium for all treatment assays. Curcumin (Lot# BCBP2315v, certified ≥95%) was isolated from C. longa , solubilized in <2.0% ethanol with 2 M HCl. EGCG (Lot# SLBQ4582v, certified ≥99%) was isolated from green tea and solubilized directly in complete medium. Regorafenib (Lot# 10589, certified ≥99%) was suspended at 1.0 mM in sterile phosphate-buffered saline (PBS). Drug treatment assays were performed in 12-well flat-bottom culture plates at a density of 1×10 5 cells/well. Dose range for EGCG was 0, 10, 50, 100, and 200 μM and curcumin was 0, 10, 50, 100, and 100 μM. Combination treatments of both compounds were with 5, 10, and 25 μM. Each single or combined treatment was incubated for 24 and 48 h. HepG2 and SW1417 cells were also treated with 2.5 μM regorafenib, alone and in combination with 10 μM EGCG + 10 μM curcumin for 48 h. All treatment assays were performed a minimum of three times followed immediately by RNA isolation for later gene expression analysis.
CELL VIABILITY AND CELL PROLIFERATION
Separate assays for viability and proliferation were plated in 12-well flat-bottom assay plates at 1×10 5 cells/well and treated with EGCG and curcumin both separately and in combination, in the previously described dose range for 24 and 48 h. Cell viability was determined by manual counts of all samples using a hemocytometer and trypan blue. Cell proliferation assays were plated in 96-well flat-bottom assay plates at 1×10 4 cells/well and were quantified using the CCK-8 Cell Proliferation Assay kit (Abcam) according to the manufacturer’s protocol. Absorbance was measured at 460 nm using a microplate reader. Viability and proliferation assays were performed a minimum of three times, and the data were combined. Statistical analysis was performed using one-way analysis of variance (ANOVA) for all treatment comparisons, followed by multiple-comparison t -test, using Prism 6 (GraphPad) software.
TOTAL RNA ISOLATION AND cDNA SYNTHESIS
HepG2 and SW1417 cell lines were first incubated for 24 h with 10 μM curcumin + 10 μM EGCG. Total RNA was isolated from all samples using the RNaqueous-Micro Total RNA Isolation Kit (Invitrogen) following the manufacturer’s protocol. The eluted RNA was quantified using NanoDrop Lite Spectrophotometer (Thermo-Scientific) and the A 260 / A 280 ratios were calculated to determine the purity of samples. All samples were within the optimal range of 2.0±0.1. Complementary DNA (cDNA) was synthesized from the isolated total RNA using Verso cDNA Synthesis Kit by Invitrogen (Thermo-Scientific) following the manufacturer’s protocol. cDNA aliquots were pooled from individual samples of each condition to be used later for quantitative polymerase chain reaction (qPCR) gene array analysis.
qPCR GENE ARRAYS
Previously prepared probes of cDNA from pooled samples of controls and the treatment condition (102 μL total) were used to assay expression of 84 genes using RT2 Profiler—Cancer Pathway Arrays (Qiagen). The RT2 arrays were run using a CFX-96 Touch Thermocycle (Bio-Rad) for 40 cycles, following Qiagen’s recommended protocol for this instrument.
qPCR VALIDATION OF TARGET GENES
Gene selection criteria for verification were based on the degree of change observed in RT2 assays, determined by Δ C t , novelty of expression, and relevance to the literature. Gene-specific primers were designed for candidate genes of interest and synthesized by IDT Technologies. All primers were tested for efficiency and amplification of a single product prior to performing validation assays.
A standard curve was constructed using pooled cDNA from all samples and run with each gene of interest in order to quantify relative expression. qPCR was performed using the following protocol: 3 min at 95°C, 40 cycles of 95°C for 15 s and 60°C for 30 s, followed by a dissociation curve. Ribosomal 18S was also quantified for all samples as the internal standard for normalization. ARNT (aryl hydrocarbon receptor nuclear translocator), ANGPT2 (angiopoietin 2), FGF2 (fibroblast growth factor 2), IGFBP7 (insulin-like growth factor–binding protein 7), and SERPINF1 (serpin family, member 1) were the candidate genes selected for validation by qPCR.
STATISTICAL ANALYSIS
One-way ANOVA was performed for all treatment comparisons, cell viability, cell proliferation, and qPCR data, followed by multiple-comparison t -test, using Prism (GraphPad) software.
RESULTS
CELL VIABILITY
In ARPE-19 control cells treated with curcumin for 24 h, there was no significant effect on viability at any concentration level. However, when the incubation was extended to 48 h, viability was significantly reduced in cells treated with 10 and 100 μM curcumin ( Figure 1A, B ). When ARPE-19 cells were subsequently treated with EGCG, viability was significantly reduced, but only at the highest concentrations: 200 μM after both 24- and 48-h exposures and in 100 μM after 48-h exposure ( Figure 1C–D ).
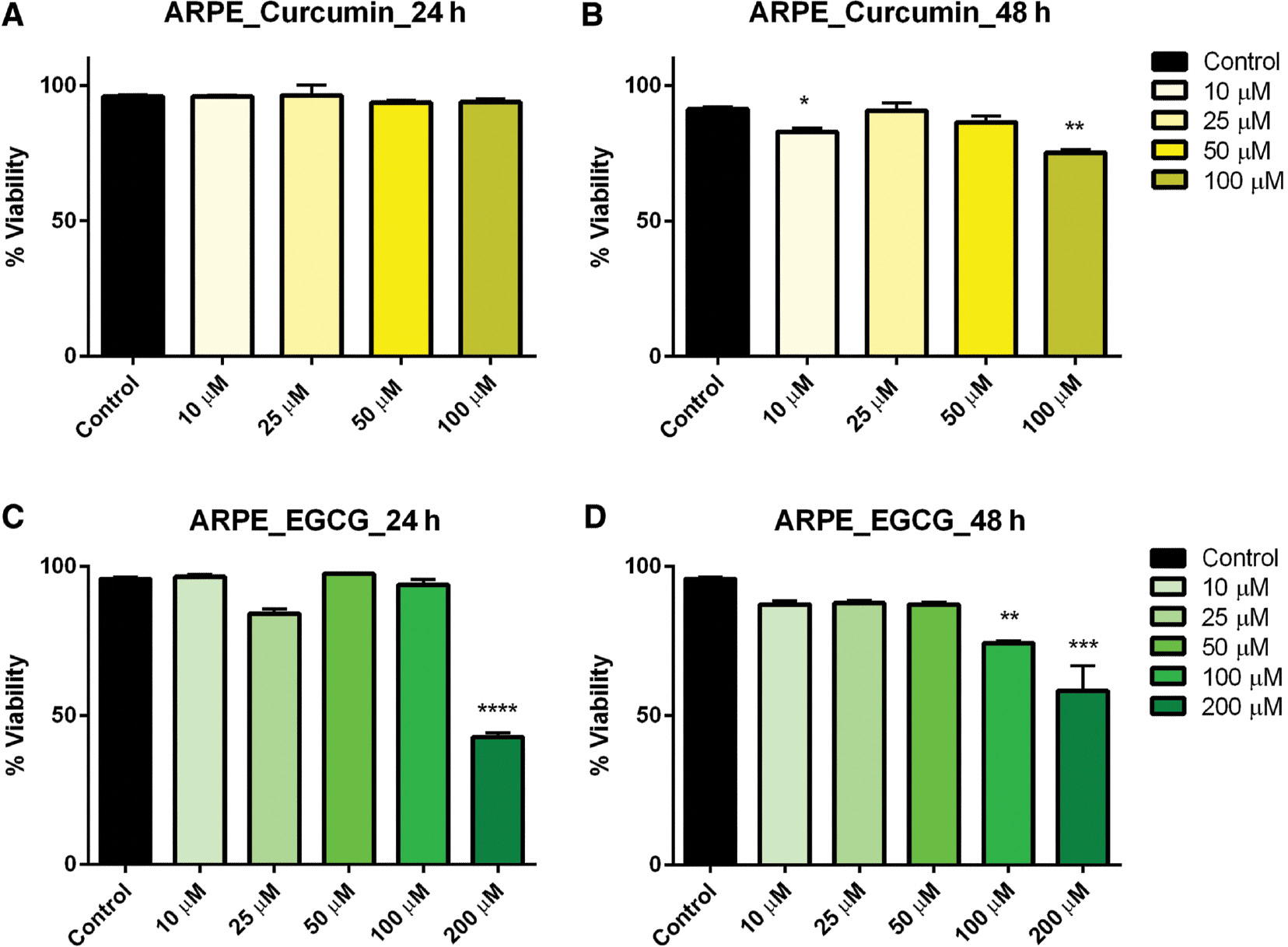
- Figure 1: -
Dose–response of curcumin and EGCG in ARPE cells at 24 and 48 h.
ARPE cells were treated with varying doses of curcumin (A, B) and EGCG (C, D) resulting in a significant effect on viability at the highest doses. Bars represent mean ± SEM ( n =3; * P <0.05, ** P <0.01, *** P <0.001, **** P <0.0001). ARPE, adult retinal pigmented epithelium; EGCG, epigallocatechin gallate; SEM, standard error of the mean.
Hepatocarcinoma (HepG2) cells treated with curcumin at varying concentrations exhibited no effect on viability after 24 h, but after 48-h exposure cell viability was significantly reduced in the 50-and 100-μM concentration ranges ( Figure 2A, B ). Similarly, EGCG at 100 and 200 μM significantly reduced the cell viability after both 24- and 48-h exposure ( Figure 2C, D ). It should be noted that there was no significant interaction between time exposure and cell viability in HepG2 cells.
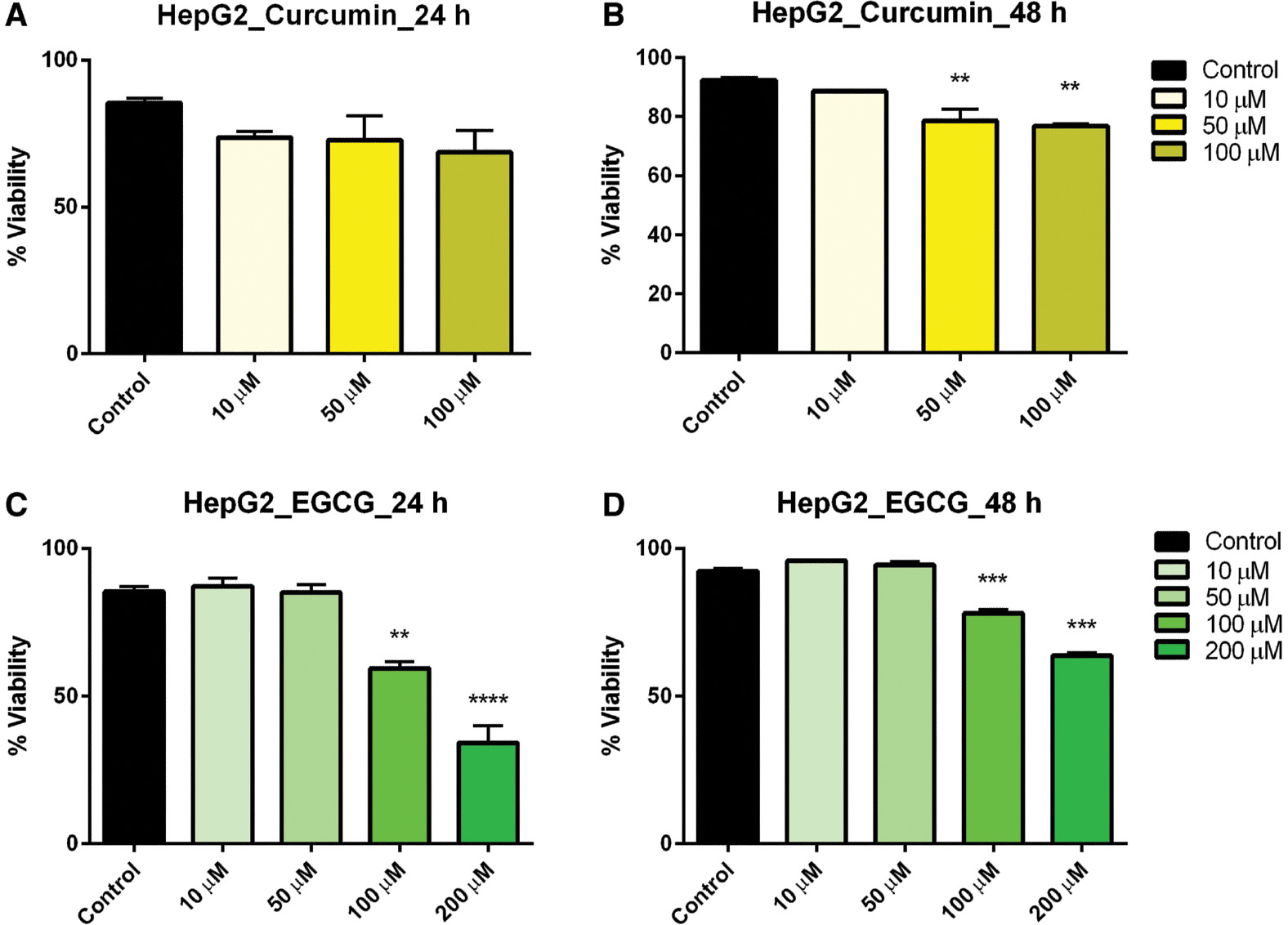
- Figure 2: -
Dose–response of curcumin and EGCG in HepG2 cells at 24 and 48 h.
HepG2 cells were treated with varying doses of curcumin (A, B) and EGCG (C, D) resulting in a significant effect on viability of HepG2 cells. Bars represent mean ± SEM ( n =3; ** P <0.01, *** P <0.001, **** P <0.0001). EGCG, epigallocatechin gallate; SEM, standard error of the mean.
Treatment within the concentration dose range of curcumin resulted in a linear reduction in viability in colorectal carcinoma (SW1417) cells after 24 h but after 48 h this effect, though significant, plateaued across all concentrations ( Figure 3A, B ). SW1417 cells exhibited mixed effects when treated with EGCG. After 24 h, a significant reduction in viability was observed in the 50-, 100-, and 200-μM concentrations; however, in the 48-h exposure, a linear decrease in viability was observed in the same concentrations, which was also significant ( Figure 3C, D ).
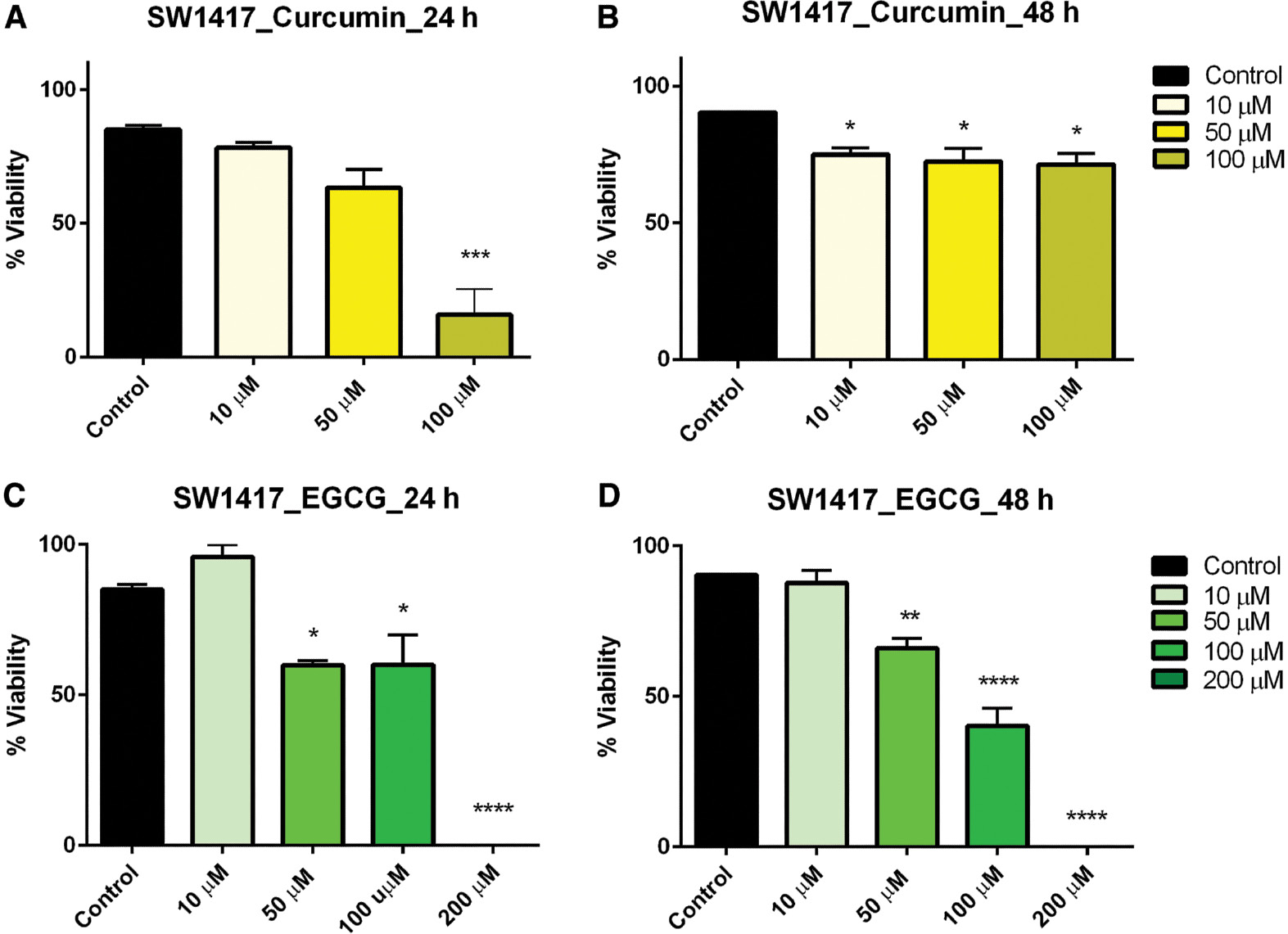
- Figure 3: -
Dose–response of curcumin and EGCG in SW1417 cells at 24 and 48 h.
SW1417 cells were treated with varying doses of curcumin (A, B) and EGCG (C, D) resulting in a significant effect on viability of SW1417 cells. Bars represent mean ± SEM ( n =3; * P <0.05, ** P <0.01, *** P <0.001, **** P <0.0001). EGCG, epigallocatechin gallate; SEM, standard error of the mean.
When curcumin and EGCG were combined in varying concentrations, we observed no significant effect on the viability of HepG2 cells after 24-h exposure ( Figure 4C ). However, when the exposure time was increased to 48 h, low-dose combinations of 5 μM curcumin + 5 μM EGCG and 10 μM curcumin + 10 μM EGCG significantly reduced viability ( Figure 4D ). Likewise, in SW1417 cells incubated with combination doses for 24 h, viability was significantly reduced in the 10 μM curcumin + 10 μM EGCG concentration ( Figure 4A ). After 48 h, viability was also significantly reduced by combinations of 5 μM curcumin + 10 μM EGCG, 10 μM curcumin + 10 μM EGCG, and 25 μM curcumin + 10 μM EGCG, suggesting that 10 μM curcumin + 10 μM EGCG had the most consistent effect on viability in both cell lines. This concentration was therefore selected for the combination treatments ( Figure 4B ).
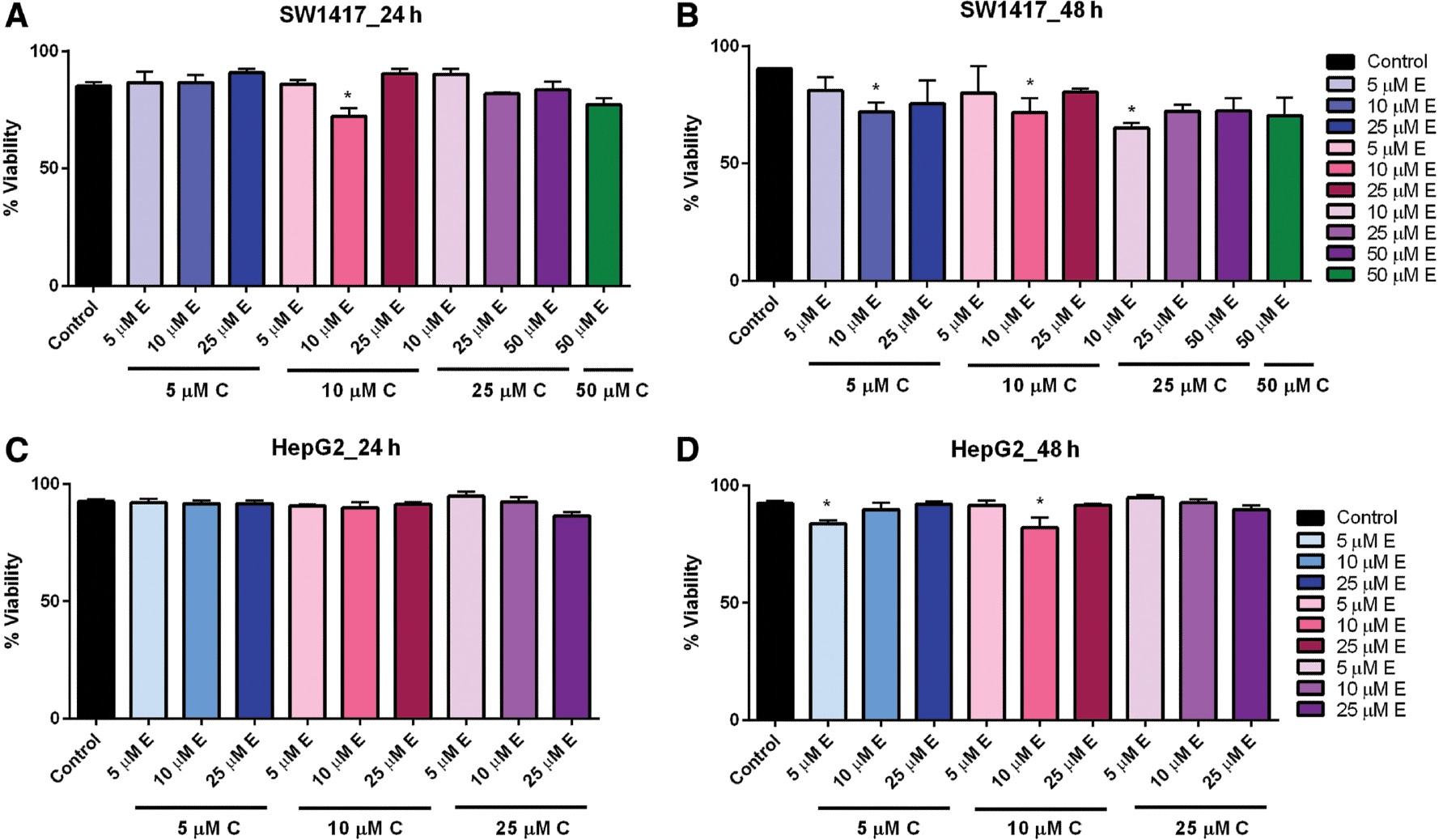
- Figure 4: -
Combination dose–response of curcumin and EGCG in HepG2 and SW1417 cells at 24 and 48 h.
SW1417 cells treated with 10 μM E + 10 μM C for 24 h (A) and 10 μM E + 5 μM C, 10 μM E + 10 μM C, and 10 μM E + 25 μM C for 48 h (B) showed a significant effect on viability. HepG2 cells treated with varying combined doses of curcumin and EGCG for 24 h (C) or 48 h (D) showed a significant effect on viability for 10 μM EGCG+10 μM curcumin. Bars represent mean±SEM ( n =3; * P <0.05). EGCG, epigallocatechin gallate; SEM, standard error of the mean.
In the control ARPE-19 cells, we observed small but significant reduced viability when cells were exposed to the combined compounds for 24 h; however, this effect was not observed after 48 h ( Figure 5A, B ). This suggests that in healthy tissue there may be an initial effect, but normally functioning cells may have the ability to recover quickly, avoiding cell death. Statistical analysis was performed by one-way ANOVA followed by multiple-comparison t -test using Prism 6 (GraphPad).
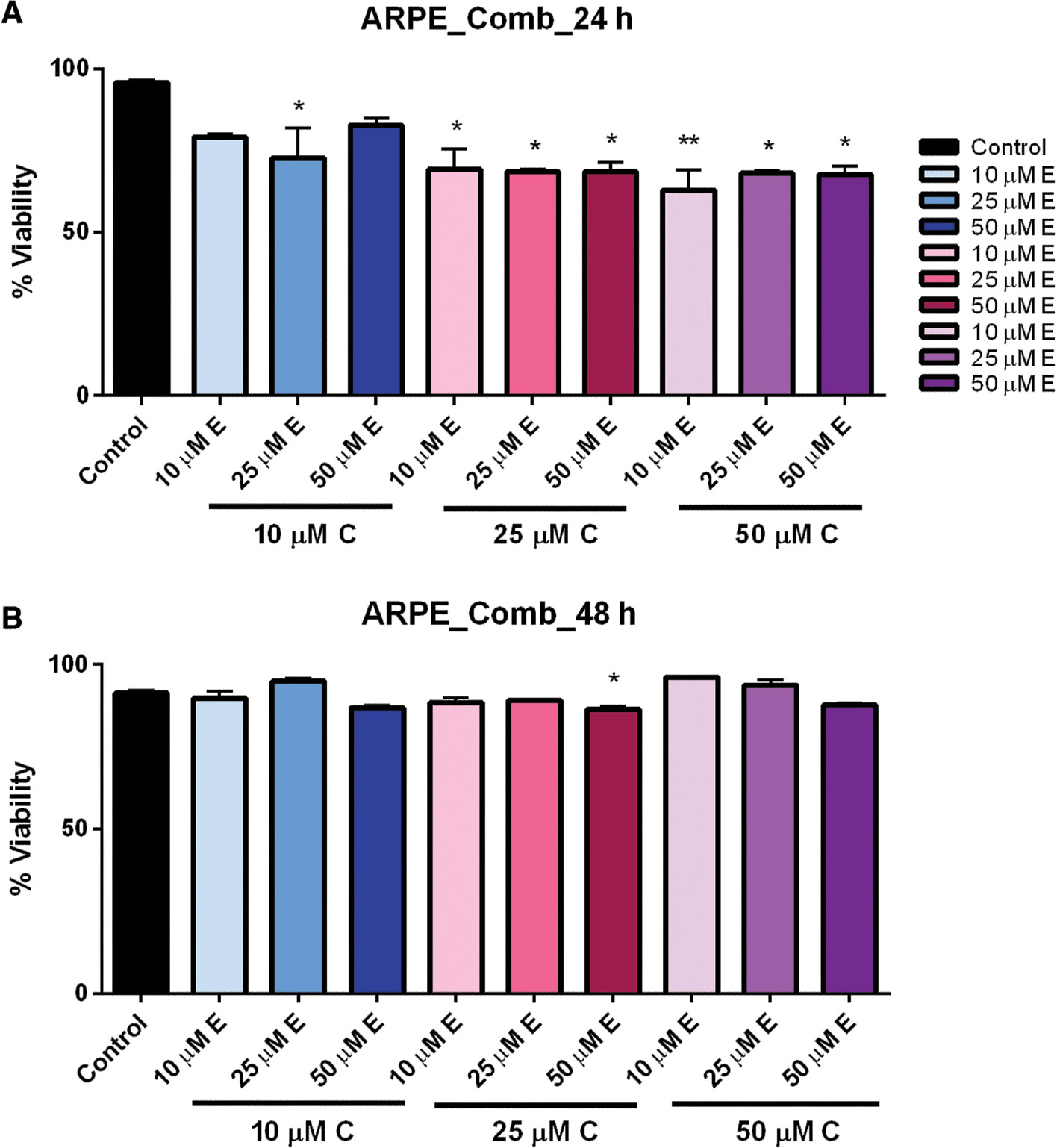
- Figure 5: -
Combination dose–response of curcumin and EGCG in ARPE cells at 24 and 48 h.
ARPE cells treated with multiple combinations of curcumin and EGCG showed significantly reduced viability at 24 h (A), but which recovered after 48 h (B). Bars represent mean ± SEM ( n =3; * P <0.05, ** P <0.01). ARPE, adult retinal pigmented epithelium; EGCG, epigallocatechin gallate; SEM, standard error of the mean.
CELL PROLIFERATION ASSAYS
The viability assays were confirmed in a time- and dose-dependent manner by monitoring cell proliferation and cell death using the CCK-8 Cell Proliferation Assay kit (Abcam). Cell proliferation was observed to decrease as concentrations of curcumin and EGCG increased in both HepG2 and SW1417 cell lines after 48 h ( Figure 6A–F ). In ARPE-19 cells, a significant decrease in proliferation was observed only in the highest doses when treated both separately with curcumin and EGCG and in combination (data not shown). The maximum effect on cell proliferation was observed in HepG2 cells treated with 100 μM curcumin ( Figure 6A ) and both HepG2 and SW1417 cells treated with 100–200 μM EGCG ( Figure 6B, E ). EGCG had the greatest overall effect of reducing proliferation in both cell lines, confirming the preliminary cell viability results.
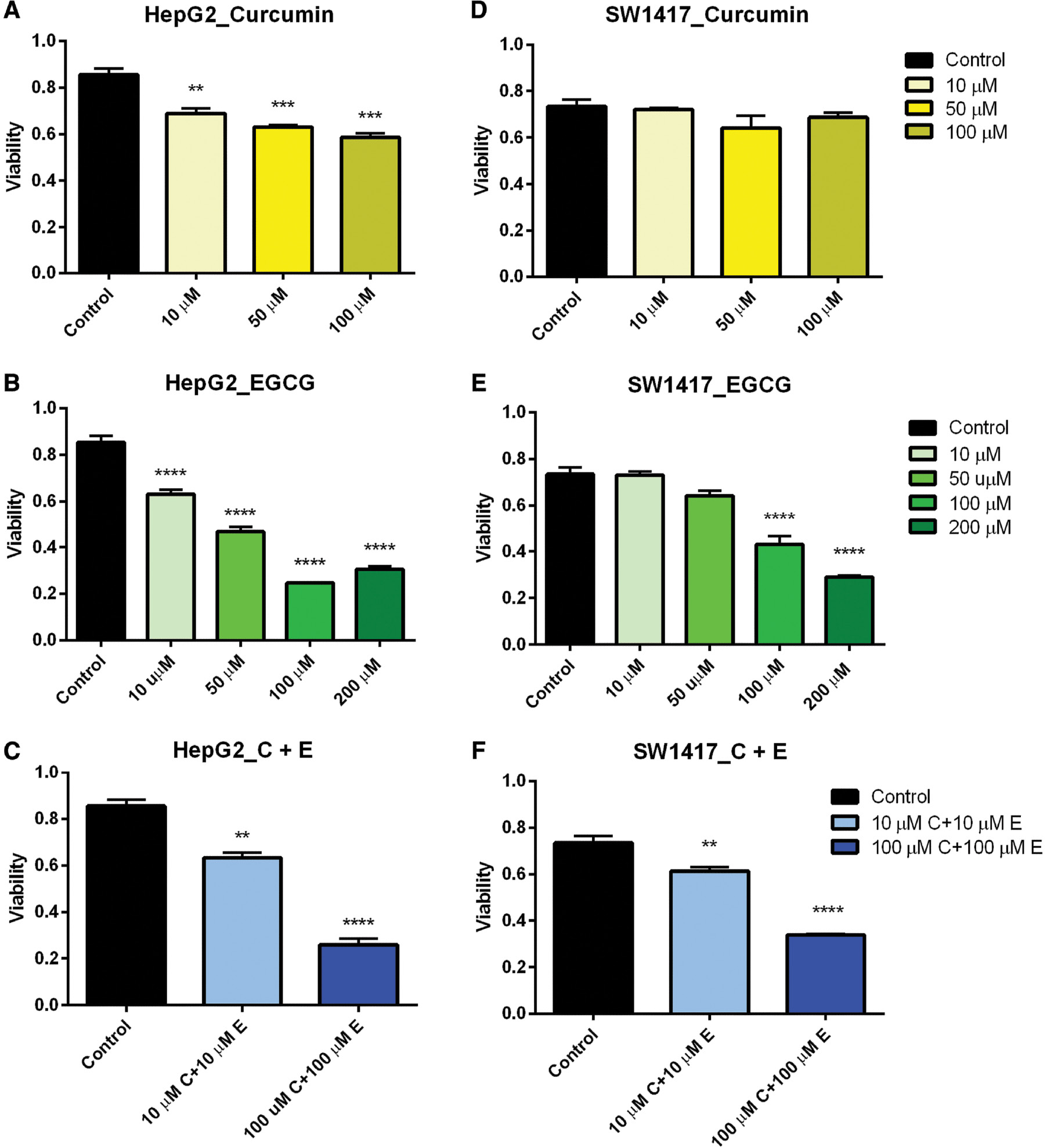
- Figure 6: -
Cell proliferation in HepG2 and SW1417 cells.
In nearly every case, proliferation was significantly inhibited when treated with a range of doses of curcumin and EGCG, both separately and in combination (A–C, E, F). In SW1417 cells treated with curcumin this effect was not significant (D). Bars represent mean±SEM ( n =3; ** P <0.01, *** P <0.001, **** P <0.0001). EGCG, epigallocatechin gallate; SEM, standard error of the mean.
When the cells were treated with a combination of 10 μM curcumin and 10 μM EGCG, ARPE-19 cells did not show any reduction in proliferation, suggesting this dose would be non-cytotoxic for therapeutic use in normal cells. However, when treated with 100 μM curcumin + 100 μM EGCG, a significant reduction in proliferation was observed, suggesting that at higher concentrations this combination could potentially be cytotoxic to healthy cells. In both SW1417 and HepG2 cell lines, proliferation was significantly decreased when treated with both the 10 μM curcumin + 10 μM EGCG and 100 μM curcumin + 100 μM EGCG combinations ( Figure 6C, F ), suggesting this drug combination may be a potential cancer therapeutic for both liver and colon cancers even at low doses.
GENE EXPRESSION ASSAY BY RT2 ARRAYS
A minimum two-fold change in gene expression was the criteria for identifying an effect of treatment. In HepG2 cells, 28 cancer genes were identified as having met the minimum change in expression as a result of treatment. Fourteen of these were downregulated relative to controls, and 14 were determined to be upregulated. In SW1417, a total of 14 genes were affected by treatment. Six of these were downregulated and eight were upregulated ( Table 1 ). The effect on gene expression in both identically treated cell lines was very distinct, suggesting that these compounds may have different mechanisms of action in different tumor types.
- Table 1: -
Effect of 10 μM curcumin+10 μM EGCG on expression of cancer targets in HepG2 and SW1417 cells.
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
| Gene | HepG2 | CCL | |
|---|---|---|---|
| Acyl-CoA synthetase long-chain fm 4 | ACSL4 | +1.23 | −0.68 |
| Apoptotic peptidase activating factor 1 | APAF1 | −0.76 | – |
| Angiopoietin 2 | ANGPT2 | – | +4.9 |
| Aryl hydrocarbon receptor nuclear translocator | ARNT | +2.59 | – |
| Aurora kinase A | AURKA | −1.23 | – |
| BCL2-like 11 | BCL2L11 | +0.99 | – |
| Baculoviral IAP repeat-containing 3 | BIRC3 | −2.52 | – |
| Carbonic anhydrase IX | CA9 | −3.26 | – |
| Caspase 7 | CASP7 | +1 | +1.68 |
| Caspase 9 | CASP9 | +0.91 | – |
| Chemokine ligand 2 | CCL2 | −1.32 | +3.59 |
| Cyclin D3 | CCND3 | +1.74 | – |
| Cadherin 2, type 1, N-cadherin (neural) | CDH2 | −2.16 | – |
| Cytochrome c oxidase 5A | COX5A | −1.23 | +0.94 |
| Dyskeratosis congenita 1, dyskerin | DKC1 | +1.07 | +2.16 |
| E2F transcription factor 4 | E2F4A | – | +1.35 |
| Excision repair cross-complementing rodent repair deficiency complementation group 3 | ERCC3 | +1.58 | +2.21 |
| V-Ets erythroblastosis virus E26 oncogene homolog 2 | ETS2 | −1.01 | – |
| Fibroblast growth factor 2 | FGF2 | −5.21 | – |
| Forkhead box C2 (MFH-1, mesenchyme forkhead 1) | FOXC2 | +1.32 | −1.02 |
| Insulin-like growth factor–binding protein 3 | IGFBP3 | −0.98 | −0.97 |
| Insulin-like growth factor–binding protein 7 | IGFBP7 | −4.31 | −1.53 |
| Keratin 14 | KRT14 | −0.97 | – |
| Polymerase beta (DNA directed) | POLB | +0.92 | – |
| Serpin peptidase inhibitor, clade F, member 1 | SERPINF1 | +2.01 | −0.97 |
| S-phase kinase-associated protein 2 (p45) | SKP2 | +0.89 | – |
| Solute carrier family 2 (facilitated glucose transporter), member 1 | SLC2A1 | −1.25 | – |
| Snail homolog 1 (Drosophila) | SNAI1 | +0.85 | – |
| Snail homolog 3 (Drosophila) | SNAI3 | – | −0.98 |
| T-box 2 | TBX2 | +0.93 | – |
| Tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase | TNKS | – | −0.9 |
| Vascular endothelial growth factor C | VEGFC | −1.09 | – |
-
EGCG, epigallocatechin gallate.
Values represent difference in threshold cycle (Δ C t ) denoting upregulation (+) or downregulation (−) in cells treated with 10 μM curcumin+10 μM EGCG relative to untreated condition.
The determination of candidate gene expression was done by selecting genes, which showed significantly more than two-fold increase or decrease when treated with a combination of 10 μM curcumin + 10 μM EGCG compared to untreated control cells. The candidate genes were further filtered based on their significant role in tumorigenesis, proliferation, and apoptosis. ARNT, FGF2, IGFBP7, and SERPINF1 were genes selected for the confirmation of expression changes in HepG2 cells; ANGPT2 and IGFBP7 were the genes selected for the confirmation of expression changes in SW1417 cells ( Table 2 ). qPCR verified gene expression in both cell lines for controls, 10 μM curcumin + 10 μM EGCG-treated (E + C), regorafenib-treated, and regorafenib combined with 10 μM curcumin + 10 μM EGCG.
- Table 2: -
Candidate genes selected for validation by qPCR in HepG2 and SW1417 cells.
- - - - - - - - - - - - -
| GenBank # | Abbreviation | Scientific name | Δ C t a |
|---|---|---|---|
| HepG2 | |||
| NM_001197325.1 | ARNT | Aryl hydrocarbon receptor nuclear translocator | +2.59 |
| NM_002006.4 | FGF2 | Fibroblast growth factor 2 | −5.21 |
| NM_001253835.1 | IGFBP7 | Insulin-like growth factor–binding protein 7 | −4.31 |
| NM_002615 | SERPINF1 | Serpin family F member 1 | +2.01 |
| SW1417 | |||
| NM_001147 | ANGPT2 | Angiopoietin 2 | +4.90 |
| NM_001253835.1 | IGFBP7 | Insulin-like growth factor–binding protein 7 | −1.53 |
-
qPCR, quantitative polymerase chain reaction.
a Values represent the difference in threshold cycle. Positive values=upregulation; negative values=downregulation.
VALIDATION OF CANDIDATE GENE EXPRESSION BY qPCR
Changes in expression of four of the five candidate genes were validated by qPCR and amplification of the target genes was confirmed by observing a single dissociation peak in all cases ( Figure 7A–F ). The same genes were then assayed for up- or downregulation when the cells were treated with 2.5 μM regorafenib separately, and in combination with 10 μM EGCG + 10 μM curcumin (E + C). The results were analyzed to determine whether regorafenib alone affected the target genes similar to the natural compounds and how expression of the genes was affected by subsequent addition of the natural compounds to regorafenib treatment.
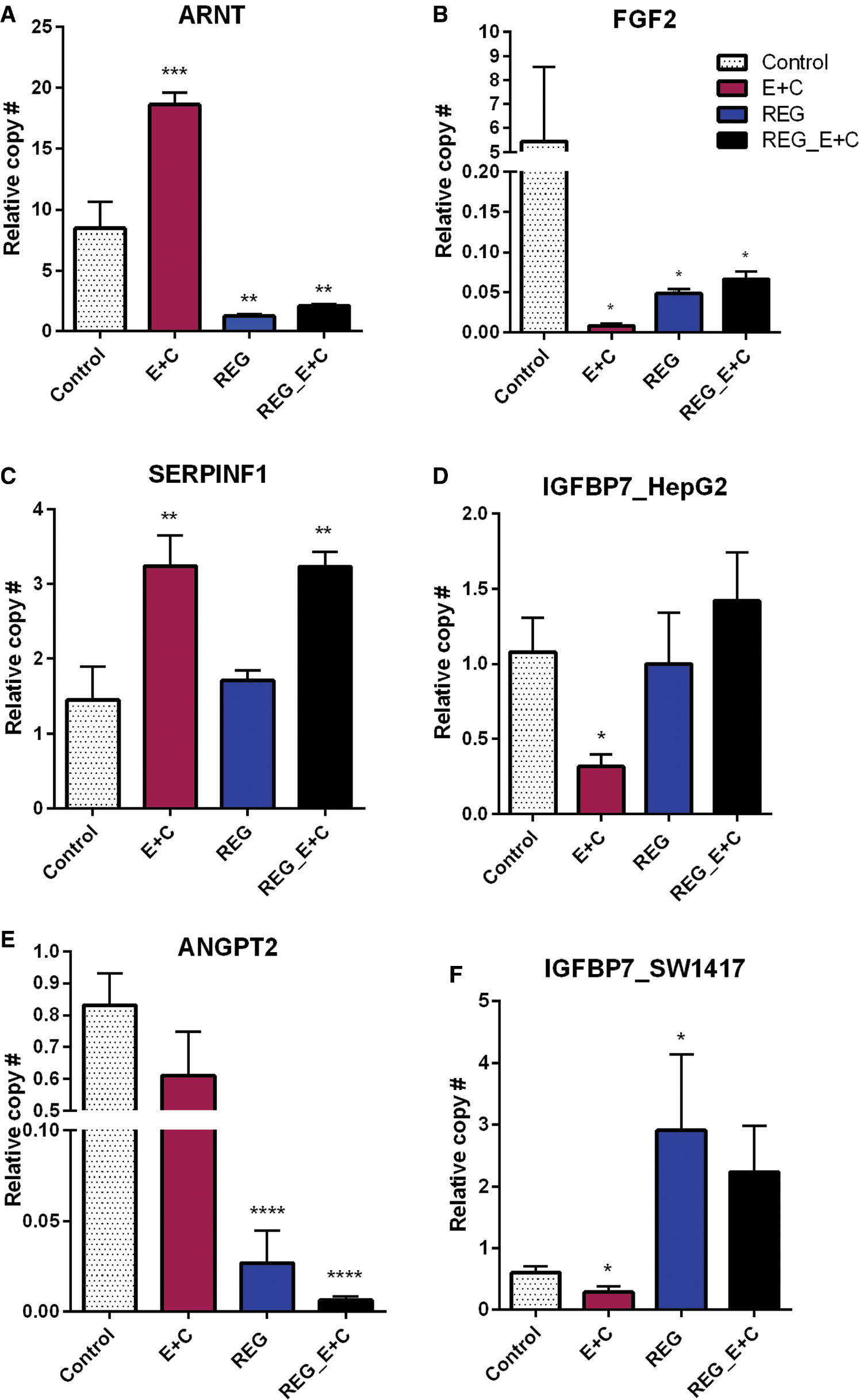
- Figure 7: -
qPCR confirmation of gene expression in HepG2 (A–D) and SW1417 (E, F) cells.
Downregulation of growth factors by E+C was confirmed (B, D, F), but regorafenib downregulated only FGF2 (B). ARNT upregulation by E+C in HepG2 cells was confirmed (A), but regorafenib had the opposite effect on this gene. SERPINF1 was upregulated by E + C but not regorafenib (C). Regorafenib combined with E+C did not reverse this effect. ANGPT2 expression was unaffected by E+C alone (E), but enhanced downregulation of regorafenib when combined with E+C. Bars represent mean ± SEM ( n =6; * P <0.05, ** P <0.01, *** P <0.001, **** P <0.0001). ARNT, aryl hydrocarbon receptor nuclear translocator; FGF2, fibroblast growth factor 2; qPCR, quantitative polymerase chain reaction; SERPINF1, serpin family, member 1.
Low-dose curcumin + EGCG downregulated expression of FGF2 and IGFBP7 in both liver and colon cancer cell lines ( Figure 7B, D, F ). Neither regorafenib alone nor in combination with 10 μM EGCG + 10 μM curcumin had this effect. ARNT was significantly upregulated by 10 μM E + 10 μM C but treatment with regorafenib resulted in a significant downregulation. Adding 10 μM E + 10 μM C to the regorafenib treatment was not able to reverse this effect ( Figure 7A ). SERPINF1 was also significantly upregulated by 10 μM E + 10 μM C, but was unaffected by regorafenib. However, when 10 μM E + 10 μM C was added to regorafenib, the same upregulation of SERPINF1 was observed ( Figure 7C ). qPCR did not confirm an effect of low-dose curcumin + EGCG on ANGPT2. However, regorafenib significantly downregulated its expression and, when combined with 10 μM E + 10 μM C, this effect was enhanced ( Figure 7E ).
DISCUSSION
The combination of curcumin and EGCG in low concentration significantly affected the survival and proliferation of HelpG2 and SW1417 cells in vitro . In much higher concentrations, these compounds had a similar effect on the normal ARPE-19 controls as well. However, when consumed in the diet under normal circumstances, the active compounds are assimilated by the body in very small amounts, thus preventing toxicity but also limiting its therapeutic potential. However, recent studies report that newer formulations of curcumin and its essential oils have demonstrated greatly enhanced bioavailability without increasing levels of cytotoxicity. 27
The regulation of important genes involved in cancer initiation and proliferation in both HCC and CRC was also highly affected. The comparative analysis of the molecular targets of curcumin combined with EGCG and regorafenib demonstrated that the low-dose curcumin and EGCG combination was significantly effective and targeted. Curcumin and EGCG induced an additive effect of regorafenib in two of the candidate genes. Hence, curcumin and EGCG may have promising applications in cancer therapy when used in combination, and an additive effect when used to supplement treatment with regorafenib. Additional studies are needed to observe the effects of higher dose combinations of curcumin and EGCG, with regorafenib and other candidate chemotherapeutics.
The ARNT is essential for normal growth and development of cells. 28 It is a transcription factor which is important for the regulation of ArH and hypoxia-inducible factor 1α (HIF-1α), both of which play a vital role in cancer proliferation. 29 ARNT expression plays an important role in tumor growth. 30 Depletion or downregulation of ARNT in normal cells can lead to abnormal angiogenesis, defective hematopoiesis, and problems related to cardiac and cranial dysfunction. 31
Metastasis results in cancer lethality, leading to an increase in malignancy and resistance to anti-cancer drugs. 32 Huang et al . demonstrated that the loss of ARNT led to a pro-metastatic phenotype, and its expression is reduced in advanced CRC. 33 This suggests that upregulation of ARNT may be significantly effective in reducing metastasis in cancer. In this study, treatment with the chemotherapy drug regorafenib resulted in downregulation of ARNT in HepG2 cells. The combination of curcumin and EGCG, when used with regorafenib, was not able to reverse this effect. Conversely, the HepG2 cells treated only with the low-dose combination of curcumin and EGCG showed significant upregulation of the gene, suggesting that it may be effective in targeting angiogenesis and inhibiting metastasis.
Serpin family 1 (SERPINF1), also known as pigment epithelium-derived factor (PEDF), is a multifunctional protein responsible for inhibiting metastasis and angiogenesis. 34 SERPINF1 plays a vital role in stimulating the maturation of the vascular environment and induction of apoptosis in cancer cells. 35 , 36 SERPINF1 can also inhibit the proliferation and migration of cancer cells that are induced by VEGF. 37 Xu et al . demonstrated that when the SERPINF1 is downregulated, the levels of thioredoxin domain-containing 5 (TXNDC5) increase. TXNDC5 is a key factor in the migration and proliferation of tumor cells, and the formation of vascular networks in endothelial cells. 38
Our data indicated a significant increase in the levels of SERPINF1 in HepG2 cells when treated with the curcumin and EGCG combination, suggesting a potential role in the reduced viability and proliferation that was observed. In contrast, regorafenib alone had no effect on the expression of SERPINF1. However, when HepG2 cells were treated with regorafenib along with curcumin + EGCG, SERPINF1 expression was upregulated to the same level as curcumin + EGCG. This finding suggests several things: (1) regorafenib does not play a role in the regulation of SERPINF1, (2) regorafenib does not inhibit the activity of low-dose curcumin + EGCG in upregulating SERPINF1, and (3) low-dose curcumin + EGCG may be a beneficial and non-cytotoxic supplement to chemotherapy by increasing anti-angiogenic support, promoting apoptosis and inhibiting metastasis. Further studies on the effect of curcumin + EGCG on SERPINF1 regulation are certainly warranted.
FGF2 belongs to the fibroblast growth factor family. Studies have shown that FGFs play a vital role in hematopoiesis, tissue regeneration, and embryonic development. In normal cells, FGF2 activity is highly regulated, and its activity is terminated via receptor internalization. FGF2 is also a prognostic marker for different types of cancers. 39 In tumor cells, FGF2 signaling is dysregulated and is one of the key promoting factors for carcinogenesis. 40 The overexpression of FGFR2 is followed by C-terminal exon retraction, leading to disrupted internalization. 41 FGF2 is known to be a potent angiogenic growth factor as well. Its overexpression induces dysregulation of FGFR1 and FGFR2, leading to increased cell proliferation, metastasis, and angiogenesis. 42 FGF2 also controls the expression of genes regulating the cell cycle, adhesion, and differentiation in endothelial cells. 43
In this study, treatment with curcumin and EGCG combined, and regorafenib alone, both resulted in significant downregulation of FGF2 in HepG2 cells. However, adding curcumin + EGCG to the regorafenib treatment did not further suppress the expression of FGF2. This demonstrates that low-dose curcumin and EGCG combination treatment may be equally effective as regorafenib in targeted therapy for FGF2 in liver cancer treatment.
It is well known that ANGPT2 plays a vital role in tumorigenesis by increasing the initiation of tumor vessel sprouts. 44 It is also known that ANGPT2 leads to the dissociation of cell–cell interaction in endothelial cells by induction of permeability. 45 In normal cells, ANGPT2 can act as a regulator for endocrine function; however, it is primarily important for the initiation of vascularization. 46 Studies have shown that levels of ANGPT2 are highly increased in tumor cells making them more susceptible to amplified angiogenesis. 47
The cancer array data indicated an unexpected increase in the expression of ANGPT2 levels in HepG2 cells treated with a combination of curcumin and EGCG. However, qPCR follow-up showed no significant difference in ANGPT2 expression between the treated cells and controls. The same cells treated with regorafenib showed a significant decrease in ANGPT2 expression, and when curcumin + EGCG was combined with regorafenib, the effect of regorafenib was enhanced. This suggests once again that low-dose curcumin + EGCG may be an effective supplement to regorafenib therapy. Due to the inconsistency of the gene expression data however, follow-up assays need to be performed to further observe the effect of these compounds on ANGPT2 expression in HepG2 cells.
IGF plays an important role in the carcinogenesis of both HCC and CRC. IGFBP7 binds to IGF, thereby blocking its activation. 48 The expression of IGFBP7 and its correlation with various tumor types is inconsistent. For example, IGFBP7 has been implicated in promoting tumor cell proliferation and angiogenesis in brain endothelial cells, 49 whereas it was anti-angiogenic and pro-apoptotic in CRC. 50 Hence, it is critical to understand the nature of a specific cancer type and the precise behavior of IGFBP7 in a particular tumor, for targeted gene therapies to be successful. In CRC, IGFBP7 can promote anchorage-independent growth in malignant cells, when it is expressed by the tumor cells themselves. 51 Conversely, other studies have shown that the loss of IGFBP7 resulted in activation of the IGF pathway leading to increased proliferation of hepatocarcinoma cells. 52
In this study, low-dose curcumin + EGCG significantly downregulated the expression of IGFBP7, suggesting it may play a role in inhibiting cell proliferation via blocking of IGFBP2. Regorafenib alone had no effect on the expression of IGFBP7 in HepG2 cells; however, when it was used in combination with curcumin and EGCG, IGFBP7 was upregulated, though not significantly. In SW1417 cells, regorafenib significantly upregulated IGFBP7, but this effect was not enhanced when regorafenib was used in combination with curcumin + EGCG. The very real but disparate effect of these treatments, both individually and combined, on IGFBP7 is not fully understood. The possibility exists that IGFBP7 plays a different role in the progression of each of these tumor types. However, the data do suggest that curcumin + EGCG may have a beneficial effect in controlling cell proliferation and inducing apoptosis in both HCC and CRC. Future studies are planned to further investigate this question.
Studies in rodents have demonstrated that poor solubility, absorption, rapid metabolism, and excretion result in only approximately 20% of ingested curcumin being actually bioavailable. 53 , 54 Efforts to enhance the clinical therapeutic effects of curcumin have employed the use of nanoparticle technology to deliver higher concentrations directly to cells rather than by oral ingestion. 55 , 56 In mouse studies however, EGCG was found to be highly soluble and have widespread uptake in numerous tissues and organs. 57 One novel approach to improving curcumin delivery is the development of metal polyphenol networks, which essentially conjugate curcumin and EGCG with iron chloride, not only making it water soluble but enhancing its efficacy against cancer cells as well. 58 The approach of using low-dose combinations of curcumin and EGCG in this study is an attempt to simulate dietary intake and bioavailable levels in humans. The hypothesis being that even the dietary-level bioavailability of these compounds may be exerting a cytoprotective effect sufficient to account in part for the discrepancy in rates of colon and liver cancers observed in different regions of India. A number of additional contributing variables would naturally account for the observed differences in cancer rates. However, the gene expression data reported here suggest that even at very low concentrations these compounds have a significant impact on important cancer pathways at the cellular level.
The benefits of dietary polyphenols extend to the gut microbiome as well. There is a growing body of evidence demonstrating that polyphenolic compounds not only promote the growth and proliferation of beneficial flora (probiotics) in the gut, but also inhibit the production of harmful microorganisms. 59 – 61 Due to its prebiotic and antioxidant activity, curcumin also supports host immune function, reducing inflammation and oxidative cell and tissue damage. 62 Cancer is well known to have multiple evasive strategies against the immune system. Like many polyphenols, the compounds used in this study are potent anti-inflammatories, which reduce the risk of collateral tissue damage, thereby potentially limiting additional chromosomal injury.
CONCLUSIONS
HepG2 and SW1417 cell lines treated with low-dose curcumin and EGCG showed significantly decreased viability and attenuated proliferation. This low-dose treatment also affected the regulation of gene targets critical for regulation of cell cycle and angiogenic processes in tumor cells. When this low-dose treatment was combined with regorafenib, it was able to enhance the effect of regorafenib on these same genes in most cases. This suggests that the addition of low-dose curcumin and EGCG to regorafenib chemotherapy may allow for reduced levels of drug administration, thus reducing the level of negative side effects as well. These data also suggest that the addition of both curcumin and EGCG (via green tea) to the average diet on a regular basis may act as a potent chemopreventive measure. Since these compounds are bio-assimilated at low levels, this would reduce the risk of cellular damage that high concentrations may potentially impose. It is also important to note that chemotherapeutics are, by definition, cytotoxic and therefore capable of imposing physiological risks, including organ failure, to the patient as well. Therefore, if phytochemical supplementation of chemotherapeutic treatment is able to enhance the efficacy of these drugs while reducing the required effective dose, patient outcomes may also be improved.
Limitations of this study include a lack of supporting data from an in vivo model, which might demonstrate the potential of phytochemicals in this treatment regimen to reverse tumor growth. In addition, further inquiry needs to be done on the role of additional gene targets which were affected by this regimen. Both of these issues are being considered for future studies. This study also lacks the use of a negative control compound. However, while there are many natural compounds and their analogs that have been the basis of modern therapeutics, there are relatively few that have been shown to be effective against cancer, particularly at the low doses used in this study. Cancer cells are masters of survival, to the point of parasitizing healthy neighboring cells, 63 which is why chemotherapeutics are necessarily so toxic by nature. One objective of this study is to accentuate the effectiveness of low doses of the selected natural compounds to target cancer cells without negatively impacting normal control cells. This may support their potential usefulness as co-therapeutics. Finally, the authors acknowledge the complexity of potential dietary, compound/compound and drug/compound interactions, which has not been addressed here and is beyond the scope and focus of this study.
References
- Heron M. Deaths: leading causes for 2017. Natl Vital Stat Rep. 2019;68:1–77.
- Kumar D, Kumar M, Saravanan C, Singh SK. Curcumin: a potential candidate for matrix metalloproteinase inhibitors. Expert Opin Ther Targets. 2012;16(10):959–72.
- Villanueva A, Newell P, Chiang DY, et al. Genomics and signaling pathways in hepatocellular carcinoma. Semin Liver Dis. 2007;27(1):55–76.
- Le Marchand L, Wilkens LR, Hankin JH, et al. A case-control study of diet and colorectal cancer in a multiethnic population in Hawaii (United States): lipids and foods of animal origin. Cancer Causes Control. 1997;8(4):637–48.
- Lin JK, Chang SC, Yang YC, Li AF. Loss of heterozygosity and DNA aneuploidy in colorectal adenocarcinoma. Ann Surg Oncol. 2003;10(9):1086–94.
- Takayama T, Miyanishi K, Hayashi T, et al. Colorectal cancer: genetics of development and metastasis. J Gastroenterol. 2006;41(3):185–92.
- Wang H, Khor TO, Shu L, et al. Plants vs. cancer: a review on natural phytochemicals in preventing and treating cancers and their druggability. Anticancer Agents Med Chem. 2012;12(10):1281–305.
- Ames BN, Gold LS, Willett WC. The causes and prevention of cancer. Proc Natl Acad Sci USA. 1995;92(12):5258–65.
- Quideau S, Deffieux D, Douat-Casassus C, Pouysegu L. Plant polyphenols: chemical properties, biological activities, and synthesis. Angew Chem Int Ed Engl. 2011;50(3):586–621.
- Shimizu M, Adachi S, Masuda M, et al. Cancer chemoprevention with green tea catechins by targeting receptor tyrosine kinases. Mol Nutr Food Res. 2011;55(6):832–43.
- Singh S, Loke YK, Furberg CD. Long-term risk of cardiovascular events with rosiglitazone: a meta-analysis. JAMA. 2007;298(10):1189–95.
- Agrawal DK, Mishra PK. Curcumin and its analogues: potential anticancer agents. Med Res Rev. 2010;30(5):818–60.
- Takeuchi T, Ishidoh T, Iijima H, et al. Structural relationship of curcumin derivatives binding to the BRCT domain of human DNA polymerase lambda. Genes Cells. 2006;11(3):223–35.
- Shin HJ, Baek KH, Jeon AH, et al. Dual roles of human BubR1, a mitotic checkpoint kinase, in the monitoring of chromosomal instability. Cancer Cell. 2003;4(6):483–97.
- Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat Rev Immunol. 2002;2(10):725–34.
- Bhattacharyya S, Mandal D, Saha B, et al. Curcumin prevents tumor-induced T cell apoptosis through Stat-5a-mediated Bcl-2 induction. J Biol Chem. 2007;282(22):15954–64.
- Prasad CP, Rath G, Mathur S, et al. Potent growth suppressive activity of curcumin in human breast cancer cells: modulation of Wnt/beta-catenin signaling. Chem Biol Interact. 2009;181(2):263–71.
- Rinaldi R, Eliasson E, Swedmark S, Morgenstern R. Reactive intermediates and the dynamics of glutathione transferases. Drug Metab Dispos. 2002;30(10):1053–8.
- Strimpakos AS, Sharma RA. Curcumin: preventive and therapeutic properties in laboratory studies and clinical trials. Antioxid Redox Signal. 2008;10(3):511–45.
- Gupta SC, Patchva S, Koh W, Aggarwal BB. Discovery of curcumin, a component of golden spice, and its miraculous biological activities. Clin Exp Pharmacol Physiol. 2012;39(3):283–99.
- Fajardo AM, Piazza GA. Chemoprevention in gastrointestinal physiology and disease. Anti-inflammatory approaches for colorectal cancer chemoprevention. Am J Physiol Gastrointest Liver Physiol. 2015;309(2):G59–70.
- Sigler K, Ruch RJ. Enhancement of gap junctional intercellular communication in tumor promotertreated cells by components of green tea. Cancer Lett. 1993;69(1):15–9.
- Gupta S, Hastak K, Afaq F, et al. Essential role of caspases in epigallocatechin-3-gallate-mediated inhibition of nuclear factor kappa B and induction of apoptosis. Oncogene. 2004;23(14):2507–22.
- Masuda M, Suzui M, Weinstein IB. Effects of epigallocatechin-3-gallate on growth, epidermal growth factor receptor signaling pathways, gene expression, and chemosensitivity in human head and neck squamous cell carcinoma cell lines. Clin Cancer Res. 2001;7(12):4220–9.
- Kissel M, Berndt S, Fiebig L, et al. Antitumor effects of regorafenib and sorafenib in preclinical models of hepatocellular carcinoma. Oncotarget. 2017;8(63):107096–108.
- Rolfo C, Bronte G, Sortino G, et al. The role of targeted therapy for gastrointestinal tumors. Expert Rev Gastroenterol Hepatol. 2014;8(8):875–85.
- Aggarwal ML, Chacko KM, Kuruvilla BT. Systematic and comprehensive investigation of the toxicity of curcuminoidessential oil complex: a bioavailable turmeric formulation. Mol Med Rep. 2016;13(1):592–604.
- Wenger RH, Gassmann M. Oxygen(es) and the hypoxiainducible factor-1. Biol Chem. 1997;378(7):609–16.
- Chandel NS, Simon MC. Hypoxia-inducible factor: roles in development, physiology, and disease. Cell Death Differ. 2008;15(4):619–20.
- Liang Y, Li WW, Yang BW, et al. Aryl hydrocarbon receptor nuclear translocator is associated with tumor growth and progression of hepatocellular carcinoma. Int J Cancer. 2012;130(8):1745–54.
- Kozak KR, Abbott B, Hankinson O. ARNT-deficient mice and placental differentiation. Dev Biol. 1997;191(2):297–305.
- Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2(1):48–58.
- Huang CR, Lee CT, Chang KY, et al. Down-regulation of ARNT promotes cancer metastasis by activating the fibronectin/integrin beta1/FAK axis. Oncotarget. 2015;6(13):11530–46.
- Rychli K, Huber K, Wojta J. Pigment epitheliumderived factor (PEDF) as a therapeutic target in cardiovascular disease. Expert Opin Ther Targets. 2009;13(11):1295–302.
- Hoshina D, Abe R, Yamagishi SI, Shimizu H. The role of PEDF in tumor growth and metastasis. Curr Mol Med. 2010;10(3):292–5.
- Wietecha MS, Krol MJ, Michalczyk ER, et al. Pigment epithelium-derived factor as a multifunctional regulator of wound healing. Am J Physiol Heart Circ Physiol. 2015;309(5):H812–26.
- Becerra SP, Notario V. The effects of PEDF on cancer biology: mechanisms of action and therapeutic potential. Nature Rev. 2013;13(4):258–71.
- Xu B, Li J, Liu X, et al. TXNDC5 is a cervical tumor susceptibility gene that stimulates cell migration, vasculogenic mimicry and angiogenesis by down-regulating SERPINF1 and TRAF1 expression. Oncotarget. 2017;8(53):91009–24.
- Korc M, Friesel RE. The role of fibroblast growth factors in tumor growth. Curr Cancer Drug Targets. 2009;9(5):639–51.
- Powers CJ, McLeskey SW, Wellstein A. Fibroblast growth factors, their receptors and signaling. Endocr Relat Cancer. 2000;7(3):165–97.
- Ahmad A, Sakr WA, Rahman KM. Novel targets for detection of cancer and their modulation by chemopreventive natural compounds. Front Bioscience (Elite Ed). 2012;4:410–25.
- Tanghetti E, Ria R, Dell’Era P, et al. Biological activity of substrate-bound basic fibroblast growth factor (FGF2): recruitment of FGF receptor-1 in endothelial cell adhesion contacts. Oncogene. 2002;21(24):3889–97.
- Dell’Era P, Coco L, Ronca R, et al. Gene expression profile in fibroblast growth factor 2-transformed endothelial cells. Oncogene. 2002;21(15):2433–40.
- Hashizume H, Falcon BL, Kuroda T, et al. Complementary actions of inhibitors of angiopoietin-2 and VEGF on tumor angiogenesis and growth. Cancer Res. 2010;70(6):2213–23.
- Scharpfenecker M, Fiedler U, Reiss Y, Augustin HG. The Tie-2 ligand angiopoietin-2 destabilizes quiescent endothelium through an internal autocrine loop mechanism. J Cell Sci. 2005;118(Pt 4):771–80.
- Mitsuhashi N, Shimizu H, Ohtsuka M, et al. Angiopoietins and Tie-2 expression in angiogenesis and proliferation of human hepatocellular carcinoma. Hepatology. 2003;37(5):1105–13.
- Zeng H, He X, Tuo QH, et al. LPS causes pericyte loss and microvascular dysfunction via disruption of Sirt3/angiopoietins/Tie-2 and HIF-2alpha/Notch3 pathways. Sci Rep. 2016;6:20931.
- Evdokimova V, Tognon CE, Benatar T, et al. IGFBP7 binds to the IGF-1 receptor and blocks its activation by insulin-like growth factors. Sci Signal. 2012;5(255):ra92.
- Pen A, Moreno MJ, Durocher Y, et al. Glioblastomasecreted factors induce IGFBP7 and angiogenesis by modulating Smad-2-dependent TGF-beta signaling. Oncogene. 2008;27(54):6834–44.
- Akiel M, Guo C, Li X, et al. IGFBP7 deletion promotes hepatocellular carcinoma. Cancer Res. 2017;77(15):4014–25.
- Rupp C, Scherzer M, Rudisch A, et al. IGFBP7, a novel tumor stroma marker, with growth-promoting effects in colon cancer through a paracrine tumor-stroma interaction. Oncogene. 2015;34(7):815–25.
- Chen D, Yoo BK, Santhekadur PK, et al. Insulin-like growth factor-binding protein-7 functions as a potential tumor suppressor in hepatocellular carcinoma. Clin Cancer Res. 2011;17(21): 6693–701.
- Anand P, Kunnumakkara AB, Newman RA, Aggarwal BB. Bioavailability of curcumin: problems and promises. Mol Pharm. 2007;4(6):807–18.
- Basu P, Maier C, Basu A. Effects of curcumin and its different formulations in preclinical and clinical studies of peripheral neuropathic and postoperative pain: a comprehensive review. Int J Mol Sci. 2021;22(9): 4666.
- Kabir MT, Rahman MH, Akter R, et al. Potential role of curcumin and its nanoformulations to treat various types of cancers. Biomolecules. 2021;11(3):392.
- Kurita T, Makino Y. Novel curcumin oral delivery systems. Anticancer Res. 2013;33(7):2807–21.
- Suganuma M, Okabe S, Sueoka N, et al. Green tea and cancer chemoprevention. Mutat Res. 1999;428(1–2):339–44.
- Chen Y, Jia D, Wang Q, et al. Promotion of the anticancer activity of curcumin based on a metalpolyphenol networks delivery system. Int J Pharm. 2021;602:120650.
- Moorthy M, Sundralingam U, Palanisamy UD. Polyphenols as prebiotics in the management of high-fat diet-induced obesity: a systematic review of animal studies. Foods. 2021;10(2):299.
- Pasinetti GM, Singh R, Westfall S, et al. The role of the gut microbiota in the metabolism of polyphenols as characterized by gnotobiotic mice. J Alzheimers Dis. 2018;63(2):409–21.
- Singh RK, Chang HW, Yan D, et al. Influence of diet on the gut microbiome and implications for human health. J Transl Med. 2017;15(1):73.
- McFadden RM, Larmonier CB, Shehab KW, et al. The role of curcumin in modulating colonic microbiota during colitis and colon cancer prevention. Inflamm Bowel Dis. 2015;21(11):2483–94.
- Saydam O, Saydam N. Deficiency of Ku induces host cell exploitation in human cancer cells. Front Cell Dev Biol. 2021;9:651818.